Notebooking & Pipelining
Advanced Demo
- This is an advanced example with lots of moving pieces, leaning into pretty idiomatic code style for
compose.mkand looking less like classicalmake. Explaining every detail isn't a priority, and some familiarity with the other demos is assumed. - The problem we're solving is itself messy, and the implementation can't be extremely pretty! The main point here is to achieve deep integration with a completely foreign tool chain in a remarkably small amount of code, and other aspects take a back seat. See instead the platform lifecycle demo for a clean and practical example on a simpler problem.
This example demonstrates how you can describe, automate, and release a highly customized jupyter lab server and pipeline, which includes custom kernels on demand and several notebooks.
As a quick overview of what we'll accomplish: the basic plan is to throw together a toolkit for formal methods. In addition to the classic notebooking experience, we want to end up with something much more extensible, while also achieving a "pipelining mode", a "tui mode", and several stand-alone tools on the way.
Other background info about this demo before we dive in:
- This example illustrates the matrioshka bridge pattern. There are other examples of this pattern, but this is one by far the most involved.
- Unlike other demos, we need several external files for this demo, including notebooks, and can't lean on tricks with embedded resources. Rather than inlining we'll use files inside the jupyter/notebooks folder, but in the end we'll still generate a frozen release that's self-contained. You can also browse the related containers/notebooks at the bottom of this page
- Unlike other demos, this example also involves a server as well as automation and pipelines i.e. the jupyter lab front-end. Side-effects like port-forwarding would usually mean the loss of statelessness that makes complex automation practical, but despite the involvement of a server / interactive frontend here we'll see several unique ways that
compose.mkstill manages to keep things fully-specified and completely reproducible, and remains remarkably amenable to automation and pipelines.
Without further delay, let's get into it.
Goals
Basic Goals
- Configuration:
- We want Jupyter lab minimally preconfigured, mostly to demonstrate the capability. See overrides.json. Including kernels and support tooling, the configuration surface for server-side jupyter is so large that beyond a simple
pip install, it actually has the characteristics of a distribution. The challenges of how to manage, encapsulate, and then automate that in a "local first" way are actually a pretty good fit forcompose.mk. - Lifecycle Management:
- We need to daemonize the front-end, but we need to be smart about it. One use-case is letting users interact with notebooks and detached-daemonization is an option, but as potential kernel developers we might want it foregrounded for logs. Notebooks and kernels can run without the webserver being live via stuff like
jupyter console, but we also might want automation over the webserver, for example withnbclient1. - Custom, containerized kernels:
- Adding/removing jupyter kernels is hard and requires a handful of files in exactly the right spot to specify the behaviour, possibly with a lot of custom code. We want to remove as much of that friction as possible at the same time as making the whole process more dynamic, basically allowing users to expose arbitrary commands in arbitrary containers as jupyter kernels directly, and without custom code.
Interface Goals
- WUI Mode:
- Also known as the standard Jupyter web interface, accessible as usual from the docker host and the browser of your choice. This of course involves port-forwarding, and we disable auth for brevity and convenience, so don't leave this running on the internet!
- TUI Mode:
- Building on the embedded TUI, we'll expose 2 dedicated content-panes: one for webserver-launch & log-tailing, and one for viewing that webserver with a full-featured browser that's console-friendly4
- Pipeline Mode:
- A serverless or completely network-sandboxed mode that's more directly amenable to automation, running all notebooks end to end.
- CLI Mode:
- Basically offering fine-grained lifecycle control and individual entrypoints for all the primitive actions that enable the above, i.e. verbs like server.stop / server.start / notebook.run / notebook.preview, etc.
Notebook Goals
- We insist on jupytext:
- Because version-control for ipynb's can be annoying, e.g. with deltas that are just timestamp-changes without output-changes, and other stuff that makes diffs a pain. Jupytext fixes this, and also lets us store notebooks-as-markdown, thus solving the problem of editing notebooks outside of jupyter. Since
jupytextalso supports bidirectional sync between raw .ipynb's and the markdown-equivalent, we will retain the ability to edit notebooks inside jupyter as well.
Choosing a Use-Case
We still need a specific use-case for our notebook project, and as mentioned above the fold, we'll try to roll out a toolkit for formal methods (hereafter simply "fmtk").
The choice here doesn't actually matter much, but it does kind of fit. Academic stuff might be more likely to be missing official docker containers, and reference material is more likely to end up with "projects" that have a significant barrier to entry, rather than shipping "tools" or "pipelines".
Ultimately, we want to expose kernels for the following tools:
- Alloy specification language
- Z3 Solver, with and without python-bindings
- Lean4 Prover, in script or theorem mode 2
We also want to accomplish more than a few specific kernels for our lab setup: we want to end up with a recipe for converting any group of containers into a group of kernels.
For the official jupyter docs about custom kernels see here and here. It can be a pretty involved process, and after it's working it still needs to be packaged/released. Let's see if we can address some of that to end up with something that feels reproducible and self-contained.
The Code
The code below satisfies all the requirements in ~150 LoC, excluding code comments, the external container definitions, the external notebooks, and other shims that jupyter requires. As mentioned earlier in the goals, this involves port-forwarding, so even though the file-system is sandboxed you probably don't want to leave it running!
What you see here is basically the necessary glue that connects everything else together. It's well-commented, but there are definitely some prerequisites! To really grok it, it's helpful to have understood the rest of the compose.mk tutorials, and already have some knowledge of jupyter and/or make. But.. don't worry much if you can't understand parts of the implementation. Again, unlike other tutorials, here we're less focused on exposition and more focused on delivering the goals. Feel free to even skip over the code here and jump directly to results / usage info.
#!/usr/bin/env -S make -f
# Demonstrates building a highly customized and self-contained console app,
# where all the application components bootstrap themselves on demand.
#
# Part of the `compose.mk` repo. This file runs as part of the test-suite.
# See the main docs: https://robot-wranglers.github.io/compose.mk/demos/notebooking
#
# USAGE: ./demos/notebooking.mk lab.tui
# USAGE: ./demos/notebooking.mk lab.pipeline
#░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░
include compose.mk
# Main entrypoint, just shows usage hints
__main__:
$(call log, \
${red}Provide a target like 'lab.tui' or 'lab.pipeline' or use 'help' for help.)
# Jupyter lab URL
lab.url.base=http://localhost:9999
lab.url=${lab.url.base}/lab/tree/notebooks
# Configuration for TUI pane contents.
lab.tui.panes=lab.notebook.open/networkx-atlas.ipynb,lab.up
# Constants to configure tmux pane geometry.
lab.tui.geometry=c3f2,231x57,0,0[231x48,0,0,1,231x8,0,49{125x8,0,49,2,105x8,126,49,4}]
export geometry?=${lab.tui.geometry}
# Jupyter constants relative to wd; lab constants
jupyter.root=demos/data/jupyter
jupyter.notebook.root=${jupyter.root}/notebooks
jupyter.kernels.root=${jupyter.root}/kernels
# Filters for jq to pull b64 image data out of jupyter notebook outputs, etc
jq.img.filter=[ .cells[] \
| select(.outputs!=null).outputs[] \
| select(.output_type=="display_data") ]
jq.imgcount.filter=${jq.img.filter} | length
# Autogenerate target scaffolding for each kernel container
$(call compose.import, file=${jupyter.root}/docker-compose.fmtk.yml namespace=fmtk)
# Autogenerate scaffolding for the jupyter-lab container.
$(call compose.import, file=${jupyter.root}/docker-compose.jupyter.yml namespace=jupyter)
## Next section uses the service scaffolding to create target-handles for language
## kernels dynamically. Although `compose.import` already created handles for all
## the containers involved, we want to plan for being able to import from other
## compose files, or use future targets as kernels directly. This means we want
## carefully designed namespaces. Kernel-invocation also involves accepting a
## filename as argument. Thus for each service, we map a new convenience target
## to an existing scaffold: `kernel.<svc>/<fname>` --> `fmtk/<svc>.command/<fname>`
##░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░
# Maps compose-services to future kernels
# Loop over the kernel list, and creating a target handle for each.
kernel_services=$(call compose.get_services, ${jupyter.root}/docker-compose.fmtk.yml)
kernel_target_names=$(foreach svc, ${kernel_services}, kernel.$(svc))
$(foreach svc, \
${kernel_services}, \
$(eval kernel.$(svc)/%:; $${make} fmtk/$(svc).command/$${*}))
# A target to list available kernels. Importantly, this is kernels according
# to compose.mk, *not* jupyter, and it is how we bootstrap the jupyter kernels.
#
# Returns both the kernels that are directly mapped from compose-services
# as well as *any targets defined in this file* matching the "kernel.*" pattern.
kernels.list:
quiet=1 \
&& ( echo "${kernel_target_names}" \
&& ${make} mk.targets.filter.parametric/kernel. ) \
| ${stream.nl.to.space}
kernel.echo/%:
@# Demonstrating a jupyter kernel that's *not* coming from the
@# fmtk compose file. This kernel simply echoes whatever code
@# is sent to it, but you can imagine it using yet another docker
@# container that we don't need to customize, or imagine it activating
@# a specific python venv, or whatever.
@#
cat ${*}
## Next section generates kernel.json files for each of the "kernel.*" targets above,
## regardless of whether those targets are static or dynamic. This step involves
## a template for jupyter kernelspec JSON, which is our starting place for generating
## dynamic kernels on the jupyter side. This works by deferring most of the kernel
## behaviour to a base class (i.e. `BaseK`), and basically adjusts environment
## variables to configure kernel-names, kernel-commands, etc.. just in time.
## For more details, see the appendix with support code/notebooks in the main docs.
##░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░
lab.gen.kernels: flux.starmap/.kernel.from.target,kernels.list
.kernel.from.target/%:; cmd="${make} ${*}" ${make} .kernel.gen/${*}
.kernel.gen/%:
mkdir -p ${jupyter.kernels.root}/${*} \
&& kfile="${jupyter.kernels.root}/${*}/kernel.json" \
&& $(call log.target.part1, generating) \
&& touch $${kfile} \
&& disp_name="`echo ${*}|cut -d. -f2`" \
&& disp_name=$${display_name:-$${disp_name}} \
&& cat ${jupyter.root}/kernel.base/kernel.json.template \
| ${jq} ".env.cmd = \"$${cmd}\"" \
| ${jq} ".env.kernel_name = \"$${disp_name}\"" \
| ${jq} ".display_name = \"$${disp_name}\"" \
| ${jq} ".language = \"$${disp_name}\"" \
| ${jq} ".env.kernel_banner = \"experimental kernel created for target @ ${*} \"" \
| ${jq} ".env.workspace = \"/lab\"" \
> $${kfile} \
&& $(call log.target.part2, ${dim_ital}$${kfile})
## Next section is a small bridge to the jupyter lab HTTP API. This isn't necessarily
## that useful since we have CLI access to jupyter.. but it shows it's accessible
## and that calls to `curl` could be replaced with `nbclient`, etc.
##░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░
api.kernels:
@# Show kernel status, according to the web ui
curl -s "${lab.url.base}/api/kernels" | ${jq} .
api.kernels.busy:
@# Shows only busy kernels
curl -s "${lab.url.base}/api/kernels" \
| ${jq} '.[] | select(.execution_state=="busy")'
api.sessions:
@# Show sessions
curl -s "${lab.url.base}/api/sessions" | ${jq} .
## Top-level interfaces for the lab.
##░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░
lab.pipeline: lab.init flux.stage.wrap/PREVIEW/lab.notebooks.preview api.kernels lab.stop
@# Pipeline-mode interface.
lab.tui: lab.init tux.open.horizontal/${lab.tui.panes}
@# UI-mode. By default this launches jupyter web
@# in one tmux pane for log viewing, then opens a
@# TUI webbrowser (carbonyl) that's pointed at it
## Top level command and control for the lab ensemble.
##░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░
lab.init: \
flux.stage.enter/INIT \
tux.require jupyter.stop jupyter.build fmtk.build \
lab.notebooks.normalize lab.gen.kernels \
lab.serve.background lab.summary \
flux.stage.exit/INIT
@# Clean initialization. This syncs updates but won't force rebuild
@# Besides background the jupyter lab server, it also synchronizes
@# raw .ipynb with paired markdown equivalent using `jupytext`.
lab.notebook.run/% lab.notebook.exec/%:
${make} lab.dispatch.quiet/self.notebook.exec/${*}
lab.notebook.preview/%:
@# Shows input and execution for a single notebook.
@# Console friendly, colored markdown rendering.
bname=`basename ${*}` \
&& label="`basename -s.ipynb ${*}`" ${make} io.draw.banner \
&& header="${dim_ital}$${bname} ${sep} ${no_ansi}" \
&& $(call log.target, $${header}( ${bold_under}input${no_ansi})) \
&& ${make} lab.dispatch.quiet/self.notebook.preview.in/${*} \
| ${stream.markdown} \
&& $(call log.target, $${header}( ${bold_under}output${no_ansi})) \
&& label="notebook finished in" ${make} flux.timer/lab.notebook.exec/${*} \
&& $(call io.mktemp) \
&& quiet=1 ${make} lab.dispatch/self.notebook.preview.out/${*} > $${tmpf} \
&& cat $${tmpf} | ${stream.markdown} \
&& error="preview failed. multiple images or incompatible file types" \
&& cat $${tmpf} | grep '!\[png\]' > /dev/null\
&& ($(call log.target, Detected image(s). Attempting preview..) \
&& ${make} lab.notebook.preview.images/${*} \
|| $(call log.target, $${error})) \
|| true
lab.notebook.imgcount/%:; cat ${*} | ${jq} -r '${jq.imgcount.filter}'
@# Return the number of images found in the given notebook
lab.notebook.preview.img/%:
$(call log.target, ${dim_cyan}Image #${bold}${cyan}$${i}\n)
$(eval index=$(shell echo $${index}))
cat ${*} \
| ${jq} -r '${jq.img.filter}[${index}].data["image/png"]' \
| base64 -d | ${stream.img}
printf '\n' > /dev/stderr
lab.notebook.preview.images/%:
@# Try to yank the images from notebook output.
@# This is naive, but we try to extract them anyway
@# for a low-resolution console preview that can
@# give a hint what changed. Unfortunately glow
@# doesn't render markdown images, but we can
count=`${make} lab.notebook.imgcount/${*}` \
&& for i in `seq 0 $$(($${count} - 1))`; \
do index=$$i ${make} lab.notebook.preview.img/${*}; \
done
lab.notebook.open/%:
@# Open the given notebook in the TUI browser.
@# (This is used as a widget by `lab.tui`)
url="${lab.url.base}/notebooks/notebooks/${*}" ${make} tux.browser
lab.notebooks:
@# List notebooks. This assumes normalization
@# has already happened, so we only return .ipynb
(ls ${jupyter.notebook.root}/*.ipynb || true) \
| grep -v Untitled | grep -v Copy
lab.notebooks.normalize normalize: lab.dispatch/self.notebooks.normalize
@# Normalize / synchronize all notebook representations.
lab.notebooks.preview: flux.starmap/lab.notebook.preview,lab.notebooks
@# Previews input, then execute notebook, displaying only output.
@# This is a pipeline, but ignores order, & has no IO across notebooks
lab.running:; strict=1 ${make} lab.ps
@# Succeeds only if the lab is currently running.
lab.serve: lab.stop lab.serve.background lab.logs
@# Blocking jupyter lab webserver
lab.serve.background: lab.up.detach
@# Non-blocking jupyter lab webserver
lab.summary: lab.wait lab.test
@# Waits for jupyterlab to ready,
@# then describes available kernels / notebooks
$(call log.target, ${dim_cyan}Available notebooks:)
${make} lab.notebooks | ${stream.indent}
$(call log.target, ${dim_cyan}Markdown notebooks:)
ls ${jupyter.notebook.root}/*.md | ${stream.indent}
lab.test: lab.dispatch/self.kernelspec.list
@# Runs a simple test-case for the lab
lab.wait: flux.loop.until/lab.running
@# Waits for the jupyter lab service to become ready
lab.webpage.open lab.wui wui: lab.wait io.browser/lab.url
@# Attempts to open a browser pointed at jupyter lab.
@# (This requires python on the host and can't run from docker)
## Low level helpers, these need to run in the lab container.
##░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░░
self.kernelspec.list:
@# Show the available kernels
$(call log.target, ${dim_cyan}Available Kernels:)
jupyter kernelspec list | sed '1d'
self.notebook.exec/%:
@# Actually run a notebook, updating it in-place.
jupyter execute ${*} --inplace
self.notebooks.normalize:
@# Sync from ipynb <--> notebooks-as-code markdown
@# bidirectionally and for each notebook.
$(call log.target, Pairing and syncing all markdown notebooks with ipynbs)
${make} self.notebook.normalize/${jupyter.notebook.root}/*.md
self.notebook.normalize/%:
@# Normalizes the given notebook
rm -f ${jupyter.notebook.root}/*.ipynb
jupytext --set-formats ipynb,md --show-changes --sync ${*} | ${stream.as.log}
self.notebook.preview.in/%:
@# Show markdown from ipnyb, pre-execution.
@# This excludes the output because it might change,
@# and also excludes code-cells when they are too large.
lines=$$(wc -l < `dirname ${*}`/`basename -s.ipynb ${*}`.md) \
&& $(call log.target, Notebook has $${lines} lines) \
&& [ "$${lines}" -lt 90 ] \
&& (\
jupyter nbconvert --to $${format:-markdown} --log-level WARN \
--stdout --MarkdownExporter.exclude_output=True ${*} ) \
|| (\
echo "*Notebook is too large; skipping input-preview*"\
&& jupyter nbconvert --to $${format:-markdown} --log-level WARN \
--stdout --MarkdownExporter.exclude_code_cell=True \
--MarkdownExporter.exclude_output=True ${*} )
self.notebook.preview.out/%:
@# Shows markdown from ipynb, post-execution.
@# (We exclude the input and labels to just show output.)
jupyter nbconvert --to $${format:-markdown} --log-level WARN \
--stdout --no-input --MarkdownExporter.exclude_markdown=True ${*}
Results & Usage
Earlier we planned for WUI Mode, a TUI Mode, a Pipeline Mode, a CLI Mode, plus frozen releases, so let's see how we did. If you've got the compose.mk repo checked out, you can interact with this demo directly as ./demos/notebooking.mk <target>. Jump directly to CLI Mode for a more complete discussion of usage, but since pipelining is maybe the most surprising and useful feature, we'll start there.
Quick links for the other topics are below:
Pipeline Mode
For Pipeline mode, use ./demos/notebooking.mk lab.pipeline. This entrypoint runs every notebook for every custom kernel in series, and in a way that's noninteractive. The output is console-friendly, and very easy to visually parse. Before the pipeline starts, all kernel-bootstrap and other setup chores are taken care of automatically. The end-result is something that's almost stateless, so it's very reproducible, but container layer-caching also means it's pretty easy to iterate on without rebuilding completely from scratch for every run.
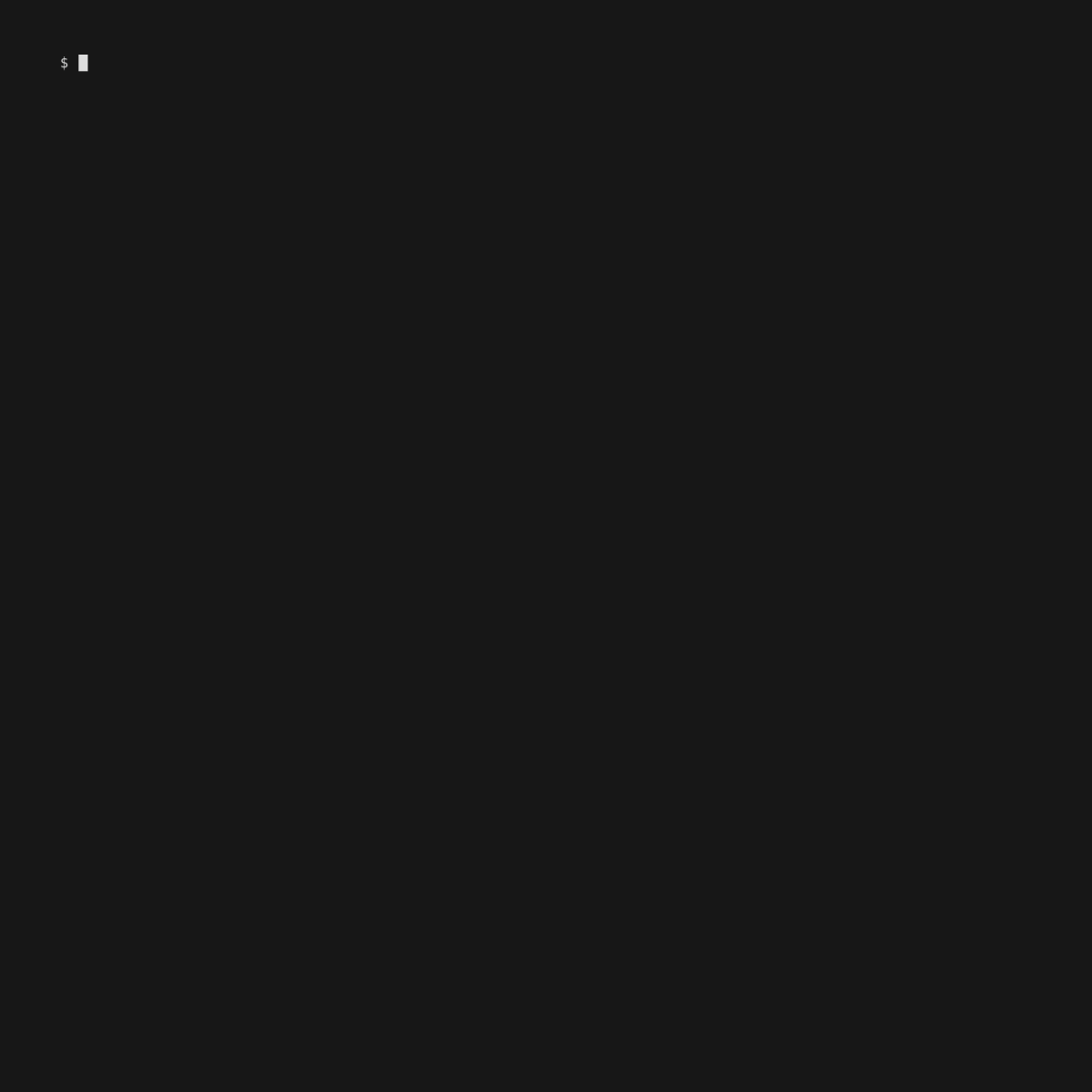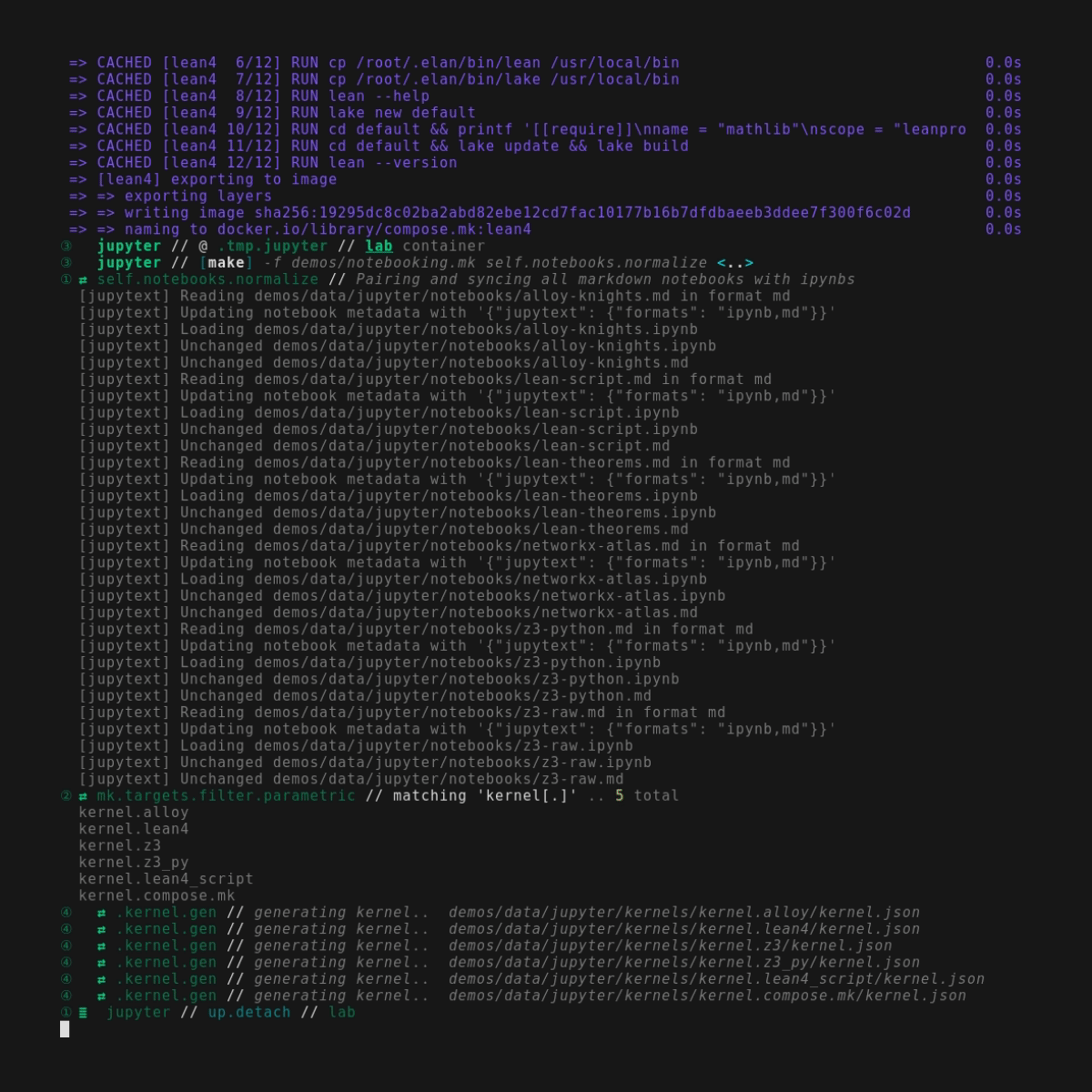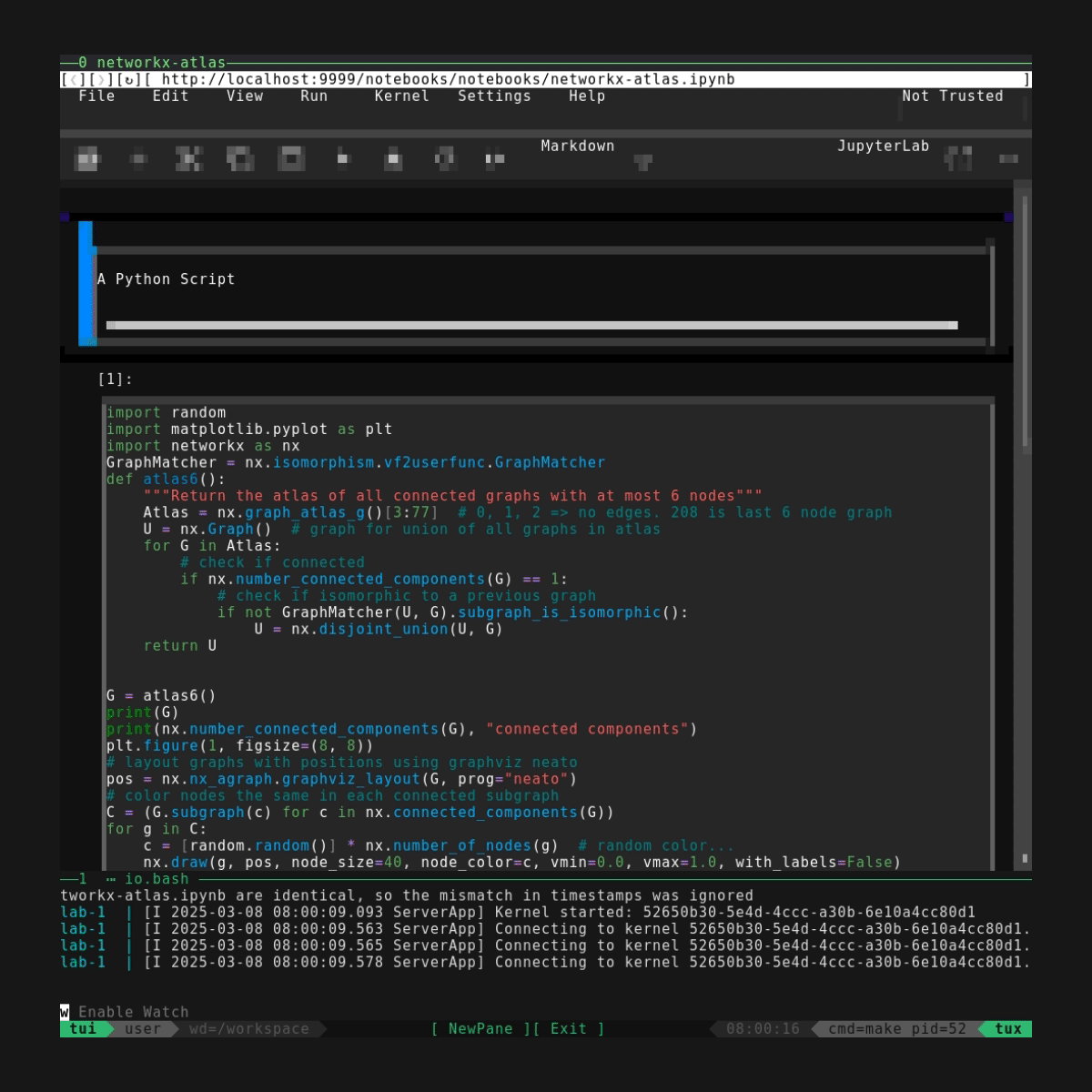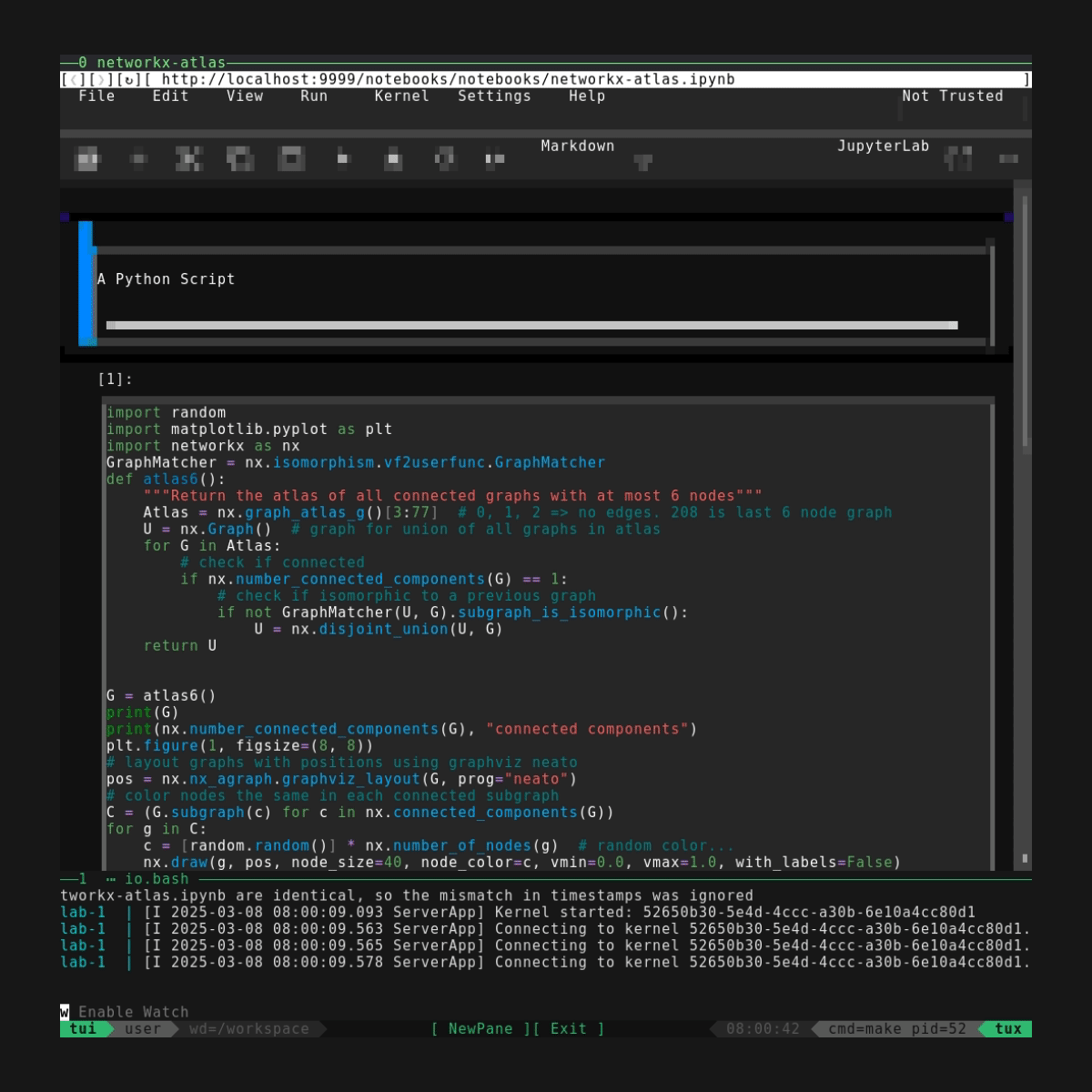
Below you can see a (very large) screenshot with a complete dump of the pipeline output. Note especially that we're not just rendering pretty markdown, but that even the more obscure languages in the codeblocks are also syntax highlighted. Notice also the pre-execution notebook synchronization with jupytext, so that version control isn't tracking a bunch of ipynb's with wiggling timestamp data. And as a bonus there's even (very low-res) image-output preview near the middle too.. this won't be a replacement for actually interactive flows, but it at least helps to figure out at a glance whether things have changed.
Pipeline mode, including syntax highlighting and image-output previews, actually works fine from Github Actions, and runs as part of the compose.mk test suite.
CLI Mode
For CLI mode, we planned for lots of granular lifecycle-management tooling, and many of these kinds of entrypoints are pretty obvious from the code.
The CLI surface is actually very large since it implicitly includes the entire static API plus scaffolded targets for our specific containers involved. For the full list of commands, try ./demos/notebooking.mk help.
However.. most users will care only about a few project-level interactions. Help is built in automatically, so to render markdown for all the local automation without the extras, just use ./demos/notebooking.mk help.local. To list only the local targets, ./demos/notebooking.mk mk.targets.
Besides the included and scaffolded targets, there's a clear and coherent project-level vocabulary emerging. For a pretty-printed version, you can use ./demos/notebooking.mk help.local.filter/lab to show all the help for the lab.* targets.
Here's a few highlights:
# Bootstraps everything, building containers, generating kernels, etc.
$ ./demos/notebooking.mk lab.init
# Start jupyter server in the foreground
$ ./demos/notebooking.mk lab.serve
# Start jupyter server in the background
$ ./demos/notebooking.mk lab.serve.background
# Attempts to open a webbrowser on the host which is pointed at the webserver.
# (Requires python, assumes the webserver is started.)
$ ./demos/notebooking.mk lab.wui
# Show running containers
$ ./demos/notebooking.mk lab.ps
# Summarize available notebooks, kernels, etc
$ ./demos/notebooking.mk lab.summary
# Show input/exec/output for all notebooks
$ ./demos/notebooking.mk lab.notebooks.preview
# Show input/exec/output for a single notebook
$ ./demos/notebooking.mk lab.notebook.preview
# Synchronize changes between .md and .ipynb notebooks
$ ./demos/notebooking.mk lab.notebooks.normalize
TUI Mode
For TUI mode, use ./demos/notebooking.mk lab.tui. We've already seen a preview since there's a video at the top of the page. As planned this starts and tails the webserver in one pane, opening a TUI browser in another. For other usage and customization details see also the general TUI docs and default keybindings info.
The TUI here is more a proof-of-concept than a truly useful feature. But one thing that's pretty interesting is that the TUI browser (as seen at the top) turns out to be surprisingly usable even for complex interactive sites like jupyter.. thanks carbonyl!
Since the carbonyl browser is just another container and can also share the network at the docker layer instead of via port-forwarding as is done here.. this raises the very intriguing possibility of using compose.mk to daemonize and visualize completely network-sandboxed interfaces to custom versions of other "runner" services like jupyter, rundeck, argo.
WUI Mode
The web UI for jupyter is mostly stock, except for add-ons created by install jupytext, annoying announcements disabled by default, etc. See the folder at demos/data/jupyter/lab for more details.
Project As Tool
For packaging, a "frozen release" type of format was promised at the top and hasn't yet been delivered. If you're interested in the details of how this works see the main docs for it here.
Skipping over the details, all we really need to do is use mk.pkg/<target>, provide an entrypoint, and ensure that the demos/ folder is archived.
$ archive=demos ./demos/notebooking.mk mk.pkg/lab.pipeline
And that's it.. now ./lab.pipeline runs as you see above, pretty much anywhere.
This is the whole project bundled into an executable archive, so now it's not dependent on working-directories or git clones, and it's easy to distribute. Initial execution pulls lots of containers, but those exist outside the archive and are naturally cached between runs. And plus.. now you can more easily hide the whole involvement of a weird tool like compose.mk ;), and yet everyone can still easily unarchive the distribution again using ./lab.pipeline --noexec --keep --target lab.unarchived.
Packaging is very practical here considering the external dependencies (compose.mk itself, plus the notebooks, plus containers for jupyter and fmtk), but again it will not package up 10gb of containerized stuff like lean+mathlib. A feature, not a bug! Despite using versioned containers/libs anywhere possible.. some are probably missed, and there are ways that pipeline output is still not reproducible if wild changes happen upstream, network connections break down, etc. What packaging really aims to fix is the typical tedious bootstrap instructions that distinguish "projects" from ready-to-go-pipelines, and prevent your "prototypes" from automatically being reusable tools.
This is convenient for an "end-to-end" test in a single command line, but there's nothing special about lab.pipeline. In a similar way, you can break off other individual entrypoints if you wish. Besides pulling data from somewhere just in time, variations on this basic theme can also include the data the pipeline runs on in addition to container specs, or be modified to work with external notebooks rather than packaged ones, and so on.
Discussion
Well now that all this exists, what exactly is it anyway? In some ways, it's easier to say what it is not:
- Obviously a TUI jupyter client cannot compete with IDE's like VSCode or fancy third-party vendors.
- Obviously a compose-backed webserver cannot compete with a k8s jupyter deployment.
On the other hand.. this whole problem space is a pecular combination of code, extensions, environments, and infrastructure. For the most part there are no visible seams, but under the hood this is orchestrating 4 containers for jupyter and the language kernels alone, and about a dozen if you include the various other utilities that we used but didn't have to install. We also completely avoided the distraction of push-pull-check loops with external repos and registries. And during the development process.. work was mostly in a single file until it was stable enough to break apart, and this further drastically reduced context-switching.
If you're interested in reproducibility, then it's not like the mere notebook code is the final word. And if you're interested in collaboration, then it turns out that it's kind of hard to simply "share" things like an IDE with a bunch of custom setup, or a kubernetes cluster with a custom deployment. A for-profit relationship with a vendor will easily get you access to an environment and handy starter-containers, but that also kind of immediately puts a fence around the work, and shelf-life on it too. While handing off a dockerhub reference for a custom container is pretty easy, that's a lot of work to commit to for mere prototyping, especially if that's outside of your interest or expertise.
Earlier the jupyter setup was compared to a "distribution", and that's really what it is, especially after you start adding default notebooks. And having hacker-friendly just-in-time kernels means you need a just-in-time distribution. Separating the code from the runtime and distributing just the notebooks works great until the runtime itself is custom or esoteric. Meanwhile for the setup we have here.. adding a new kernel is as simple as adding a new container specification, and afterwards it is attached automatically.
In the end the main features for this demo are the packaging, the dynamic kernels, and the approach to pipelining. There's an aspect of this that's related to "local first" development, but it also frees you up to run jupyter outside of jupyter, abstract away the details of "tasks in containers", and maybe avoid using a vendor by just treating this whole pipeline as a single step in another platform like Airflow / Argo. As mentioned previously.. the whole pipeline also works fine from Github Actions
This is just a demo and not a real project, but if you're thinking it's not production ready.. you can of course ship the flexible base kernel shim to pypi, and the kind of additional automation that ships the fmtk containers to dockerhub is practically a one-liner with compose.mk. Doing both of those things just reduces the support code though, and won't answer the majority of the goals!5
One final thought before the support-code appendices. The main thing this example accomplishes is a small but powerful framework for extending jupyter lab. But it's also perhaps the first nontrivial demonstration of an extension for compose.mk itself. In various other demos we've already seen examples of how you can use compose.mk to build a bridge across the host/container gap, and to expose and mix components from foreign tools and foreign languages.
Now that we've effectively created a recipe for mapping containers -> targets -> kernels, we have essentially extended the default homoiconicity of compose.mk in a new and powerful way. In principle.. we could choose to throw away the webserver and our naive end-to-end-pipeline, and then start scripting with notebooks as our "first-class components" of choice, leaving compose.mk primitives to handle things like IO and control-flow between components. Whether or not that's really useful it is certainly kind of interesting, and we're 95% of the way there without even trying.
Appendix: Support Code
Appendix: Main Jupyter Container
name: jupyter
services:
# main jupyter container, includes the server and the main python kernel
lab:
hostname: lab
image: compose.mk:lab
entrypoint: jupyter
ports: ['9999:9999']
build:
context: .
dockerfile_inline: |
FROM compose.mk:dind_base
USER root
RUN apt-get install -qq -y python3-venv python3-pip
RUN pip3 install -q jupyter==1.1.1 nbconvert==7.16.6 ipywidgets==8.1.5 jupytext==1.16.7 --break-system-packages
RUN apt-get install -qq -y graphviz graphviz-dev
RUN pip install networkx==3.4.2 matplotlib==3.10.1 pygraphviz==1.14 --break-system-packages
# NB: paths are relative to the docker-compose file!
COPY lab/ /usr/local/share/jupyter/lab/
command: >-
lab --allow-root --no-browser
--ip=0.0.0.0 --port=9999 --ServerApp.token=
--ServerApp.password=
--ServerApp.root_dir=/lab/demos/data/jupyter/
environment:
DOCKER_HOST_WORKSPACE: ${DOCKER_HOST_WORKSPACE:-${PWD}}
CMK_DIND: "1"
workspace: /lab
working_dir: /lab
volumes:
- ./kernels/:/usr/share/jupyter/kernels/
- ./kernel.base/:/usr/share/jupyter/kernel.base/
- ${DOCKER_HOST_WORKSPACE:-${PWD}}:/lab
- ${DOCKER_SOCKET:-/var/run/docker.sock}:/var/run/docker.sock
Appendix: FMTK Containers
Below you can see the definitions for all the containers involved the formal-methods toolkit. Due to size constraints, TLA+ integration is left as an exercise to the reader but here's a hint^[3] =)
# default to line-nos: https://github.com/jupyterlab/jupyterlab/issues/2395
# docker images https://github.com/Z3Prover/z3/discussions/5740
# https://github.com/z3prover/z3/pkgs/container/z3
# docker pull ghcr.io/z3prover/z3:ubuntu-20.04-bare-z3-sha-d66609e
name: notebooking
services:
# Only used for inheritance, this collects common config.
abstract: &abstract
image: hello-world
stdin_open: false
tty: false
environment:
DOCKER_HOST_WORKSPACE: ${DOCKER_HOST_WORKSPACE:-${PWD}}
CMK_DIND: "1"
workspace: /lab
working_dir: /lab
volumes:
- ${DOCKER_HOST_WORKSPACE:-${PWD}}:/lab
- ${DOCKER_SOCKET:-/var/run/docker.sock}:/var/run/docker.sock
# https://github.com/Z3Prover/z3/discussions/5740
# z3 official container does not include bindings
z3: &z3
<<: *abstract
hostname: z3
image: compose.mk:z3
build:
context: .
dockerfile_inline: |
FROM ghcr.io/z3prover/z3:ubuntu-20.04-bare-z3-sha-d66609e
RUN apt-get update -qq && apt-get install -qq -y python3 python3-pip
RUN pip3 install z3-solver==4.14.0.0 -q
z3_py:
<<: *z3
depends_on: ['z3']
hostname: z3_py
image: compose.mk:z3_py
entrypoint: python
# no official container for alloy-lang,
# but this one can run headless and return JSON
alloy:
<<: *abstract
hostname: alloy
image: compose.mk:alloy
build:
context: .
dockerfile_inline: |
FROM eloengineering/alloy-cli:4.2
RUN apt-get update && apt-get install -y make procps g++ openjdk-17-jdk-headless
entrypoint: alloy-run
command: /dev/stdin
# Reuse the lean container we built in `demos/lean.mk`
lean4: &lean4
<<: *abstract
hostname: lean4
image: compose.mk:lean4
entrypoint: lean
build:
context: .
dockerfile_inline: |
FROM debian:bookworm
SHELL ["/bin/bash", "-x", "-c"]
RUN apt-get update -qq && apt-get install -qq -y git make procps curl sudo
RUN curl https://raw.githubusercontent.com/leanprover/elan/master/elan-init.sh -sSf \
> /usr/local/bin/elan-init.sh
RUN bash /usr/local/bin/elan-init.sh -y
RUN cp /root/.elan/bin/lean /usr/local/bin
RUN cp /root/.elan/bin/lake /usr/local/bin
ENV PATH="$PATH:/root/.elan/bin/"
RUN lean --help
RUN lake new default
RUN cd default && \
printf '[[require]]\nname = "mathlib"\nscope = "leanprover-community"' >> lakefile.toml
RUN cd default && lake update && lake build
RUN lean --version
lean4_script:
<<: *lean4
depends_on: ['lean4']
hostname: lean4_script
image: compose.mk:lean4.script
entrypoint: ['lean','--run']
Appendix: Jupyter Kernel Shims
We need a base kernel to extend, so this is a version of the official jupyter example that defers to the environment for most of it's configuration. With this in place, we only need to override a few key values to use a compose.mk task (or anything else really). See the usage of .kernel.json.template in the main code for more details.
import os
import subprocess
import tempfile
from ipykernel.kernelbase import Kernel
class BaseK(Kernel):
implementation = "Container"
implementation_version = "1.0"
language = "no-op"
language_version = "0.1"
language_info = {
"name": "Any text",
"mimetype": "text/plain",
"file_extension": ".txt",
}
banner = "Experimental BaseK kernel"
def do_execute(
self, code, silent, store_history=True, user_expressions=None, allow_stdin=False
):
"""
Execute the content of the Jupyter cell, write it to a file,
and then run the file using a subprocess, returning the output.
"""
workspace = os.environ.get("workspace", ".")
os.chdir(workspace)
with tempfile.NamedTemporaryFile(dir=None, prefix=".tmp.ipy.cmk") as tmpfile:
fname = os.path.basename(tmpfile.name)
self.log.debug(f"tmpfile: {fname}")
try:
with open(fname, "w") as f:
f.write(code)
cmd = os.environ.get("cmd", "cat .")
cmd = f"{cmd}/{fname}"
self.log.warning(f"starting subprocess: {cmd}")
result = subprocess.run(cmd, shell=True, capture_output=True, text=True)
output = result.stdout
stderr = result.stderr
if result.returncode != 0:
output += stderr
if not silent:
resp = (
self.iopub_socket,
"stream",
{"name": "stdout", "text": output},
)
self.send_response(*resp)
if result.returncode != 0:
self.log.critical(f"stderr: {stderr}")
return {
"status": "error",
"traceback": [],
"ename": "failed",
"execution_count": self.execution_count,
"evalue": f"{result.returncode}",
}
else:
os.remove(fname)
return {
"status": "ok",
"user_expressions": {},
"payload": [],
"execution_count": self.execution_count,
}
except Exception as e:
resp = "stream", {"name": "stderr", "text": str(e)}
if not silent:
self.send_response(self.iopub_socket, *resp)
return {
"status": "error",
"evalue": str(e),
"execution_count": self.execution_count,
"ename": type(e).__name__,
"traceback": [],
}
if __name__ == "__main__":
from ipykernel.kernelapp import IPKernelApp
IPKernelApp.launch_instance(kernel_class=BaseK)
And the template JSON used with overrides for each of the individual kernels:
{
"argv": [
"python",
"-c",
"import os; from basek import BaseK; from ipykernel.kernelapp import IPKernelApp; banner=os.environ.get('kernel_banner','kernel_banner'); name=os.environ.get('kernel_name','kernel_name'); IPKernelApp.launch_instance(kernel_class=type(name,(BaseK,),dict(banner=banner,)))",
"-f",
"{connection_file}"
],
"env": {
"PYTHONPATH": "/usr/share/jupyter/kernel.base/"
}
}
Appendix: Jupytext Notebooks
The notebooks being used to exercise the kernels are nothing very fancy, but since we use jupytext and notebooks-as-code instead of notebooks-as-JSON.. there is the benefit that we can render most of them below directly. You can also see all the individual notebooks at demos/data/jupyter/notebooks.
If your own vibes are everywhere continuous but nowhere differentiable, then you might enjoy some demos of the Weierstrass function. The "vibes coding" notebook @ demos/data/jupyter/notebooks/vibes.md is AI-generated, breaks with our main "formal methods toolkit" theme, and is ommitted from inclusion below for brevity. The purpose of that notebook is to generate some test-images with standard matplotlib for checking out the results of rendering them using pipeline mode.
Demo: Alloy Specification Language
See also:
- Knights puzzle: https://www.hillelwayne.com/post/knights-knaves/
- Headless alloy analyzer that returns JSON: docker-alloy-cli
abstract sig Person {}
sig Knight extends Person {}
sig Knave extends Person {}
pred Puzzle {
#Person = 2
some disj A, B :Person {
A in Knight <=> (A + B) in Knave
}
}
run Puzzle for 10
Demo: Lean4 Script
def main : IO Unit := IO.println "Hello, world!"
Demo: Lean Theorems
Law of the excluded middle in lean4.
See also:
Note: success is quiet here
section
open Classical
example : A ∨ ¬ A := by
apply byContradiction
intro (h1 : ¬ (A ∨ ¬ A))
have h2 : ¬ A := by
intro (h3 : A)
have h4 : A ∨ ¬ A := Or.inl h3
show False
exact h1 h4
have h5 : A ∨ ¬ A := Or.inr h2
show False
exact h1 h5
end
Demo: Python & NetworkX
See also:
- https://networkx.org/documentation/stable/auto_examples/graphviz_layout/plot_atlas.html
import random
import matplotlib.pyplot as plt
import networkx as nx
GraphMatcher = nx.isomorphism.vf2userfunc.GraphMatcher
def atlas6():
"""Return the atlas of all connected graphs with at most 6 nodes"""
Atlas = nx.graph_atlas_g()[3:77] # 0, 1, 2 => no edges. 208 is last 6 node graph
U = nx.Graph() # graph for union of all graphs in atlas
for G in Atlas:
# check if connected
if nx.number_connected_components(G) == 1:
# check if isomorphic to a previous graph
if not GraphMatcher(U, G).subgraph_is_isomorphic():
U = nx.disjoint_union(U, G)
return U
G = atlas6()
print(G)
print(nx.number_connected_components(G), "connected components")
plt.figure(1, figsize=(8, 8))
# layout graphs with positions using graphviz neato
pos = nx.nx_agraph.graphviz_layout(G, prog="neato")
# color nodes the same in each connected subgraph
C = (G.subgraph(c) for c in nx.connected_components(G))
for g in C:
c = [random.random()] * nx.number_of_nodes(g) # random color...
nx.draw(g, pos, node_size=40, node_color=c, vmin=0.0, vmax=1.0, with_labels=False)
plt.show()
Demo: Z3 with Python Bindings
Examples from:
from z3 import *
Object = DeclareSort("Object")
Human = Function("Human", Object, BoolSort())
Mortal = Function("Mortal", Object, BoolSort())
# a well known philosopher
socrates = Const("socrates", Object)
# free variables used in forall must be declared Const in python
x = Const("x", Object)
axioms = [ForAll([x], Implies(Human(x), Mortal(x))), Human(socrates)]
s = Solver()
s.add(axioms)
# prints sat so axioms are coherent
print(s.check())
# classical refutation
s.add(Not(Mortal(socrates)))
# prints unsat so socrates is Mortal
print(s.check())
from z3 import *
# 8 queens
# We know each queen must be in a different row.
# So, we represent each queen by a single integer: the column position
Q = [Int(f"Q_{row + 1}") for row in range(8)]
# Each queen is in a column {1, ... 8 }
val_c = [And(1 <= Q[row], Q[row] <= 8) for row in range(8)]
# At most one queen per column
col_c = [Distinct(Q)]
# Diagonal constraint
diag_c = [
If(i == j, True, And(Q[i] - Q[j] != i - j, Q[i] - Q[j] != j - i))
for i in range(8)
for j in range(i)
]
solve(val_c + col_c + diag_c)
Demo: Z3 Raw
See also: * De Morgans law proof from Wikipedia
Note: *Success here is 'unsat'
(declare-fun a () Bool)
(declare-fun b () Bool)
(assert (not (= (not (and a b)) (or (not a)(not b)))))
(check-sat)
References
-
See the official nbclient docs ↩
-
We already got a head start with a lean container in an earlier demo ↩
-
Actually fleshing out the formal-methods toolkit with many more containers is tempting, but this demo is a limited-space proof of concept so that it can actually run end-to-end in github actions. ↩
-
If you're a jupyter expert and know a better way to get custom kernels in a dynamic way, please do create an issue describing it so this demo can be slimmed and hardened. ↩